Machine Learning
Decision Tree Algorithms
Roadmap
What We’ll Cover Today
What decision trees refer to in the world of supervised learning
Different types of decision tree models
- Single decision tree estimators
- Decision tree ensembles—e.g., random forests and gradient boosting
Some key terms or concepts we should know before fitting decision tree algorithms—inclusive of leaves, pruning and bagging
How to grow random forests in
Python
What Are Decision Trees?
Decision Trees in the Wild
To put it simply, decision trees are tree-like flowcharts that connect decisions to potential outcomes—i.e., by visualizing the pathways that link our choices to results in the face of uncertainty, chance events etc.
We can think about decision trees as a series of conditional (if/else) statements:
- Creating coding rules for ethnographic data.
- Deciding whether to wear shirt \(x\) or \(y\) before heading outside to face the world.
The Tennis Example
Are we going to play tennis today? Let’s think through the different possibilities.
If there is a fair bit of cloud cover (i.e., overcast conditions), we’re playing tennis.
If it’s sunny, we’ll play tennis—but only if it’s \(\le\) 27.5o C with humidity.
If it’s raining, we’ll play tennis—but only if it’s not windy.
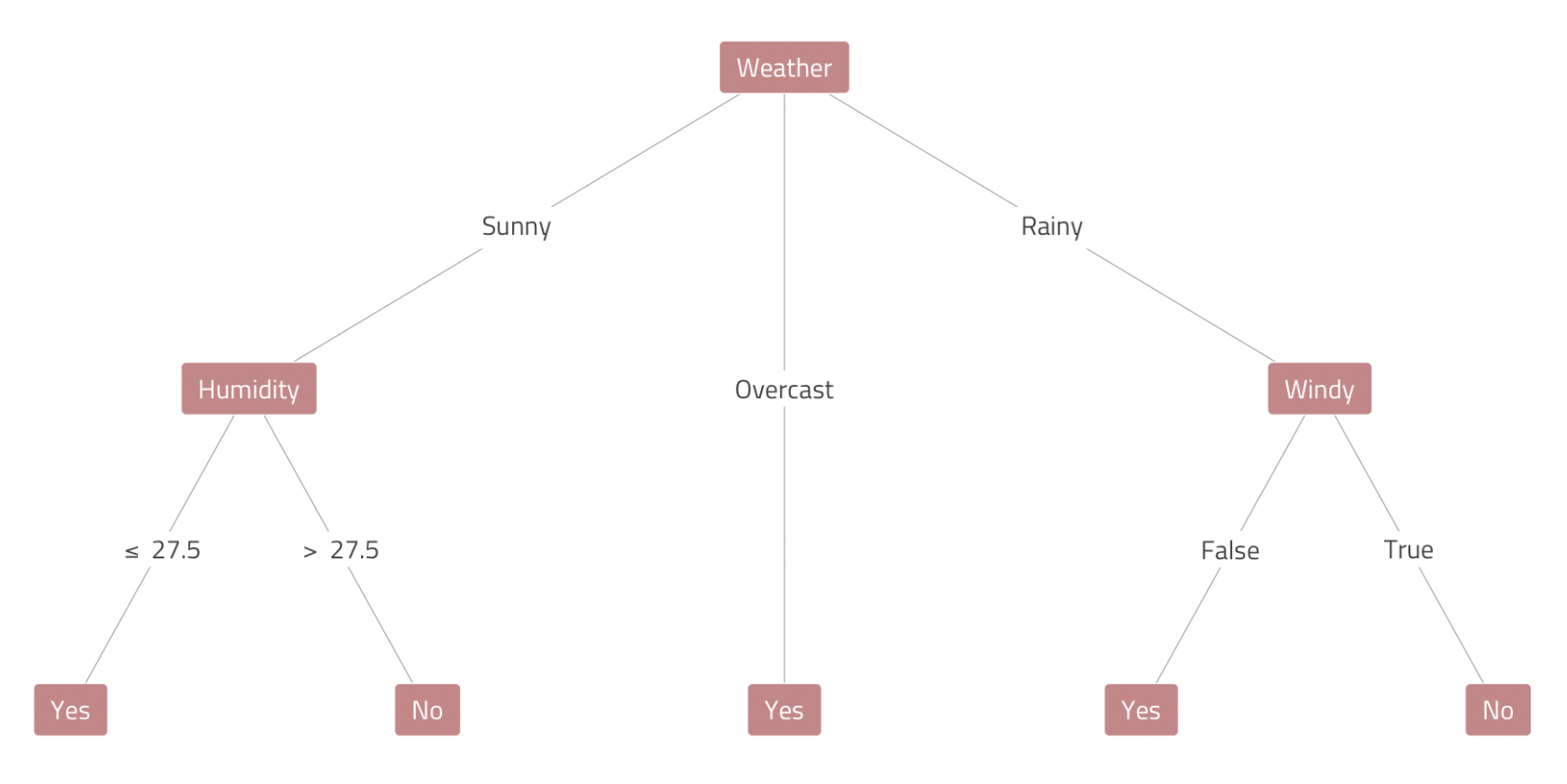
Decision Trees in Machine Learning

In the world of SML, tree-based algorithms apply the logic of decision trees to resolve regression and classification problems.
To make predictions, decision tree models take an unseen data point on a journey trough the feature space—or pathways that can lead to different endpoints (terminal nodes) depending on the observation’s features.
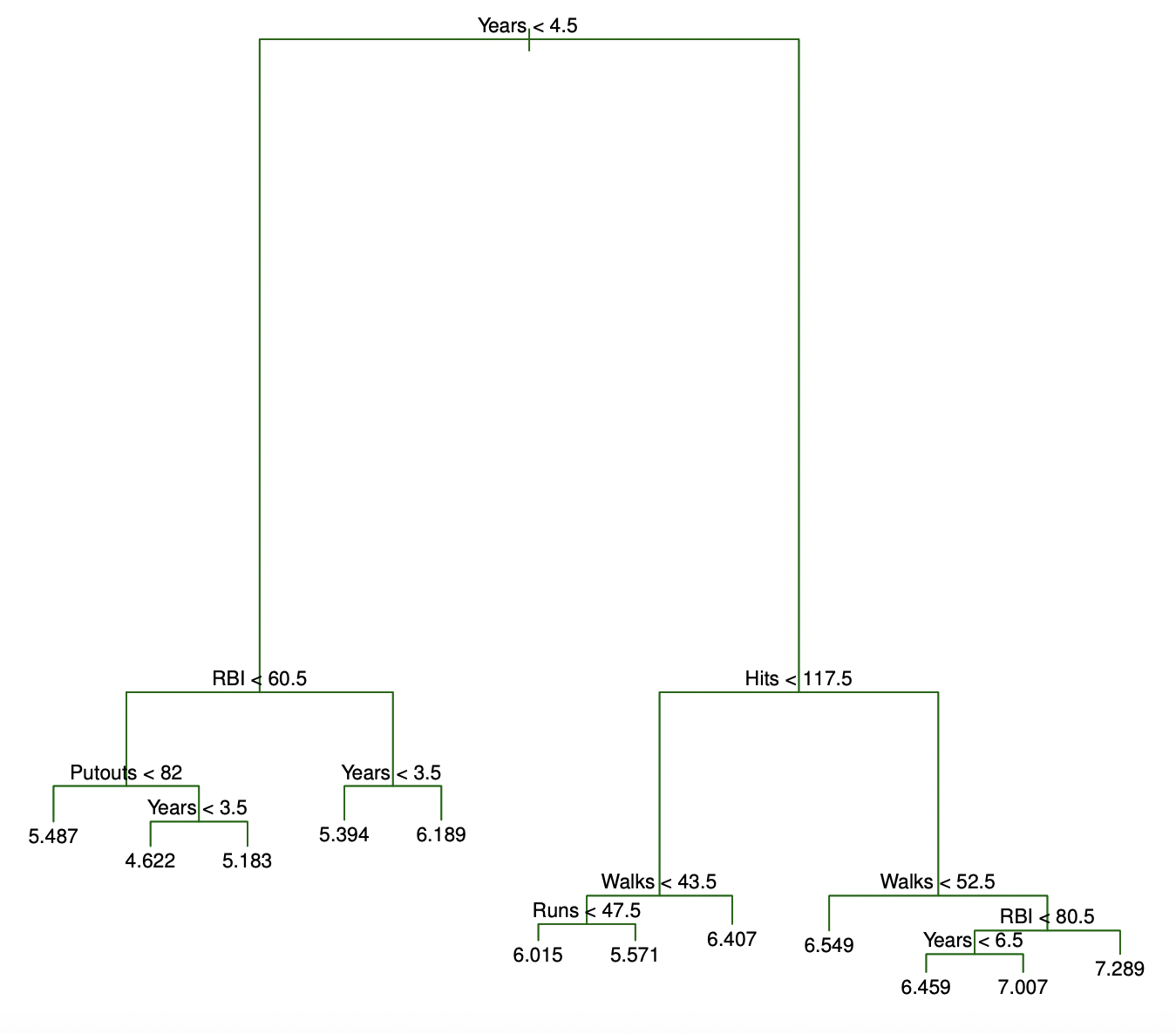
To generate the tree diagram on the left—and explore decision tree estimators for the first time in
Python—click here.
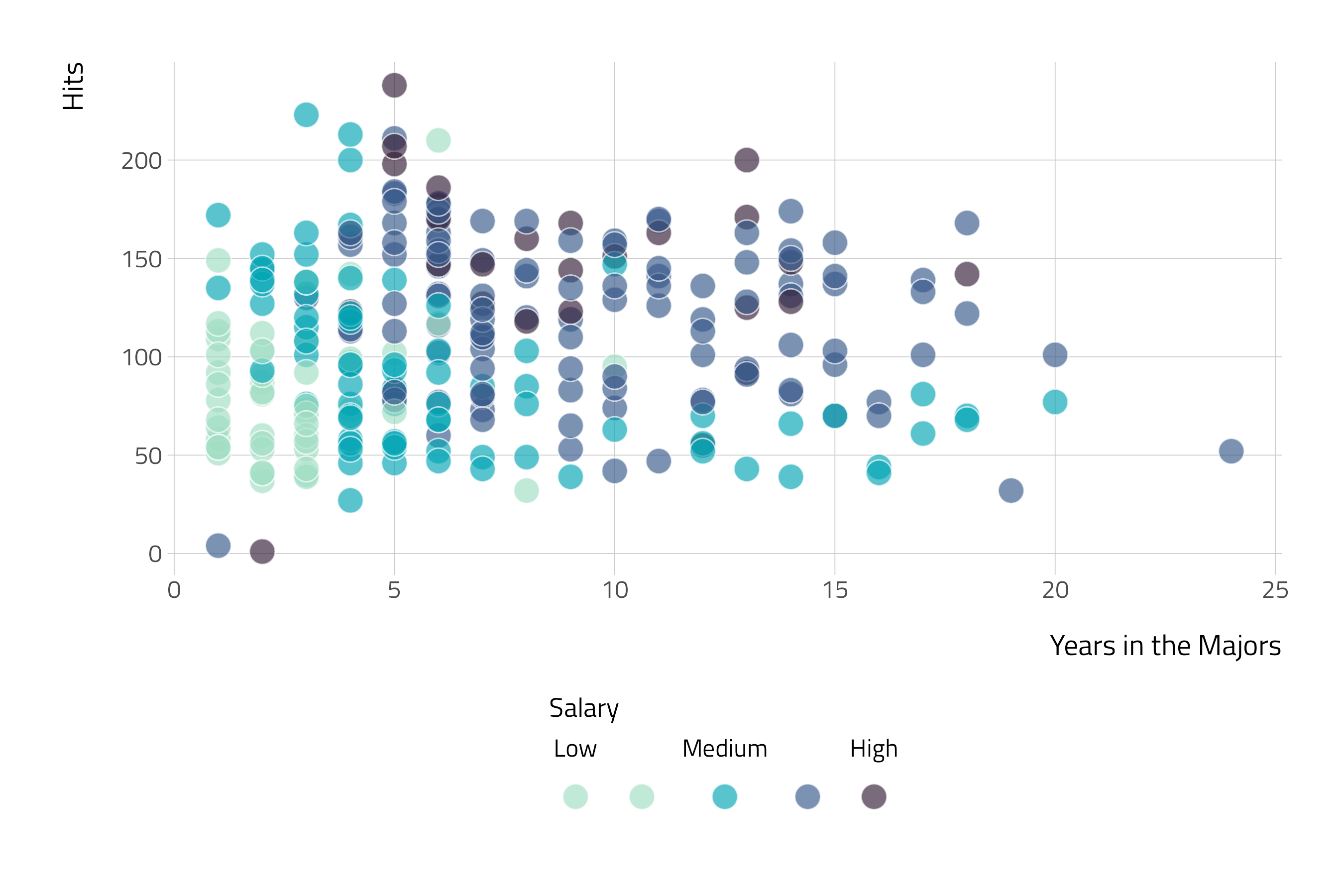
The Baseball Example
The following example comes from Introduction to Statistical Learning.
Objective
Use a decision tree to predict a baseball player’s salary using data from ’86.
Data
# A tibble: 322 × 21
Player AtBat Hits HmRun Runs RBI Walks Years CAtBat CHits CHmRun CRuns
<chr> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
1 -Andy Al… 293 66 1 30 29 14 1 293 66 1 30
2 -Alan As… 315 81 7 24 38 39 14 3449 835 69 321
3 -Alvin D… 479 130 18 66 72 76 3 1624 457 63 224
4 -Andre D… 496 141 20 65 78 37 11 5628 1575 225 828
5 -Andres … 321 87 10 39 42 30 2 396 101 12 48
6 -Alfredo… 594 169 4 74 51 35 11 4408 1133 19 501
7 -Al Newm… 185 37 1 23 8 21 2 214 42 1 30
8 -Argenis… 298 73 0 24 24 7 3 509 108 0 41
9 -Andres … 323 81 6 26 32 8 2 341 86 6 32
10 -Andre T… 401 92 17 49 66 65 13 5206 1332 253 784
# ℹ 312 more rows
# ℹ 9 more variables: CRBI <int>, CWalks <int>, League <fct>, Division <fct>,
# PutOuts <int>, Assists <int>, Errors <int>, Salary <dbl>, NewLeague <fct>The Baseball Example (cont.)
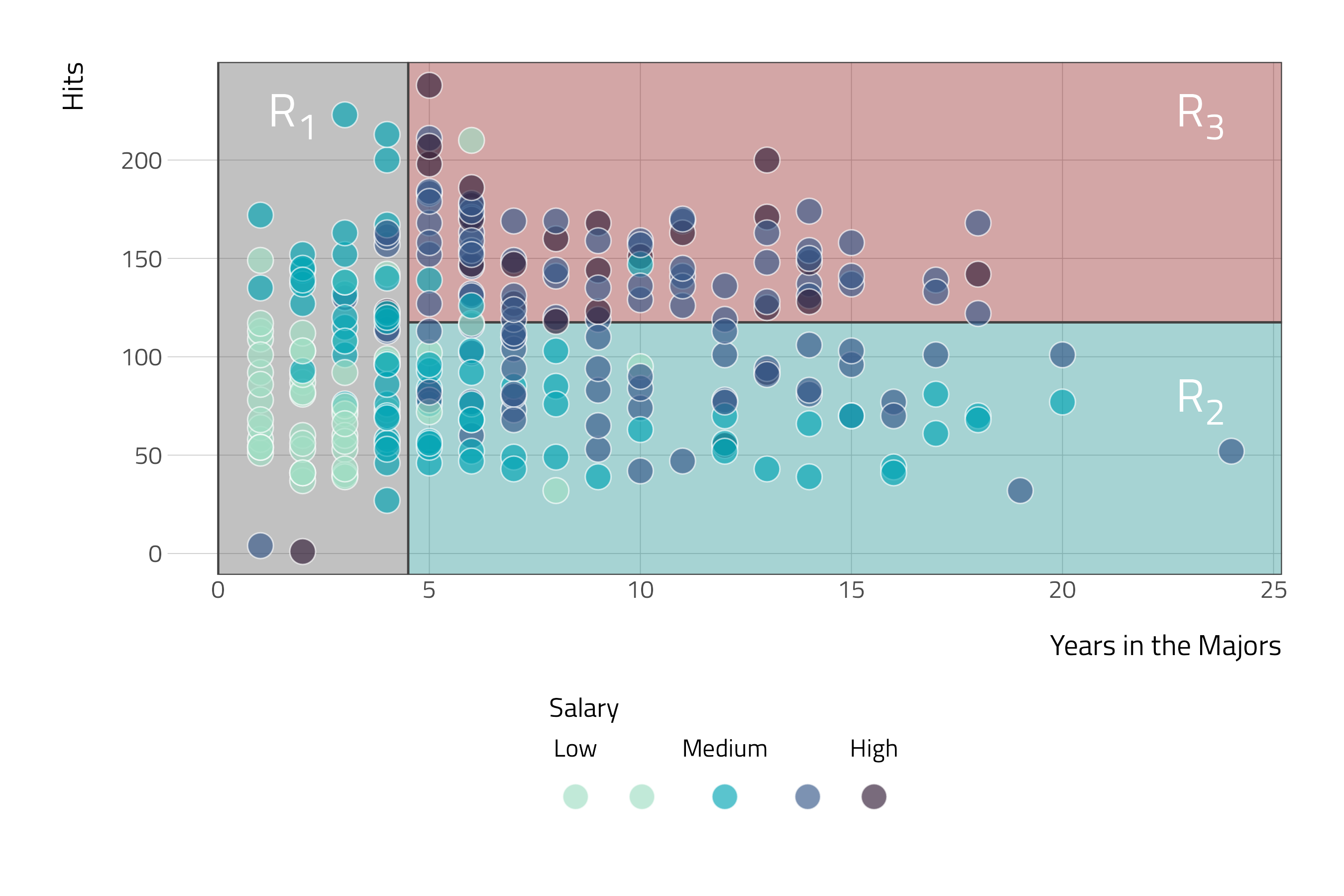
Here, the feature space is being stratified or segmented into three distinct regions:
- \(R_1\) = {\(X\) |
Years\(<\) 4.5} - \(R_2\) = {\(X\) |
Years\(\ge\) 4.5,Hits\(<\) 117.5} - \(R_3\) = {\(X\) |
Years\(\ge\) 4.5,Hits\(\ge\) 117.5}
- \(R_1\) = {\(X\) |
The Baseball Example (cont.)
| Player | Years | Hits | salary |
|---|---|---|---|
| 8 | 160 |
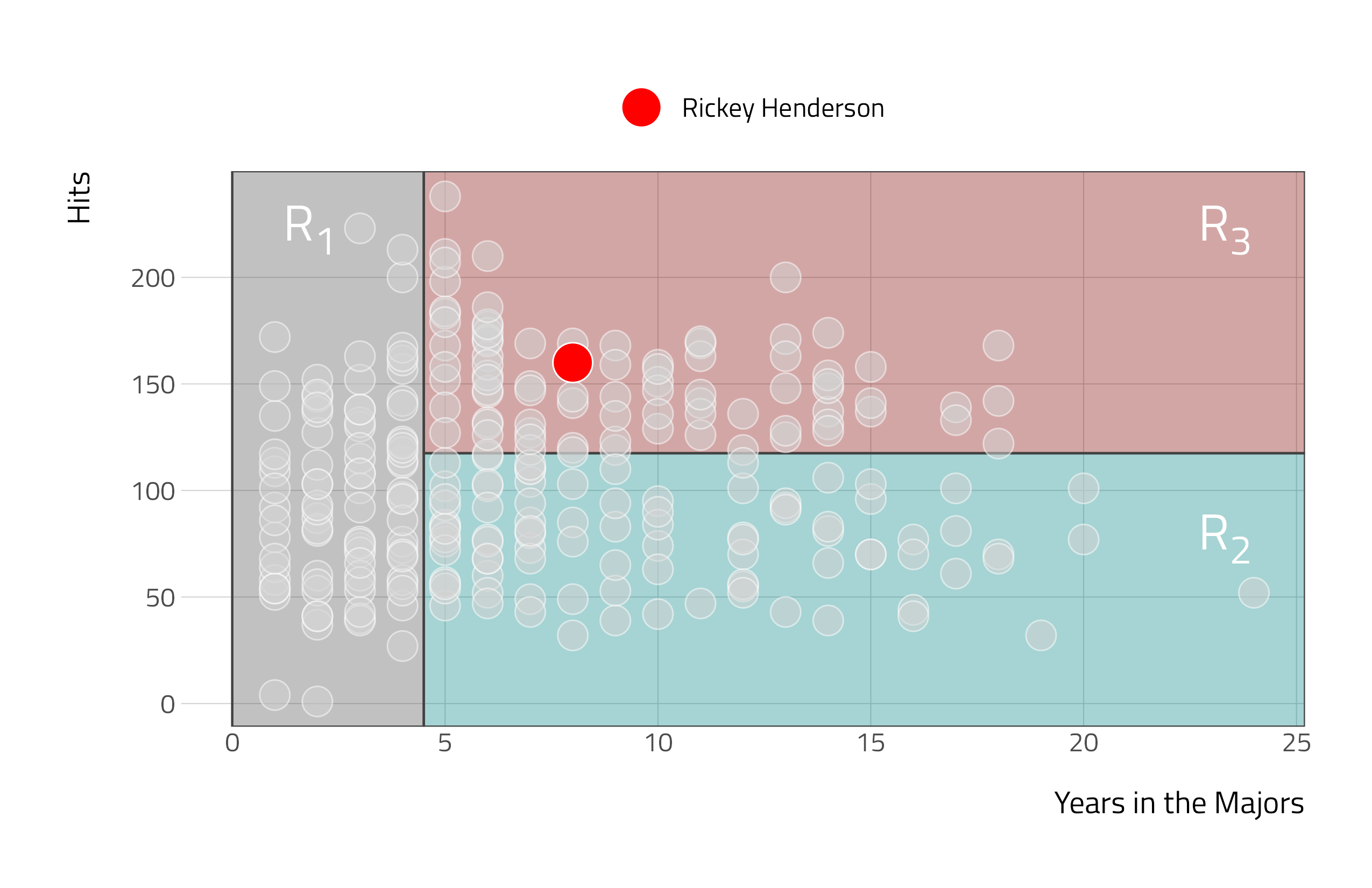
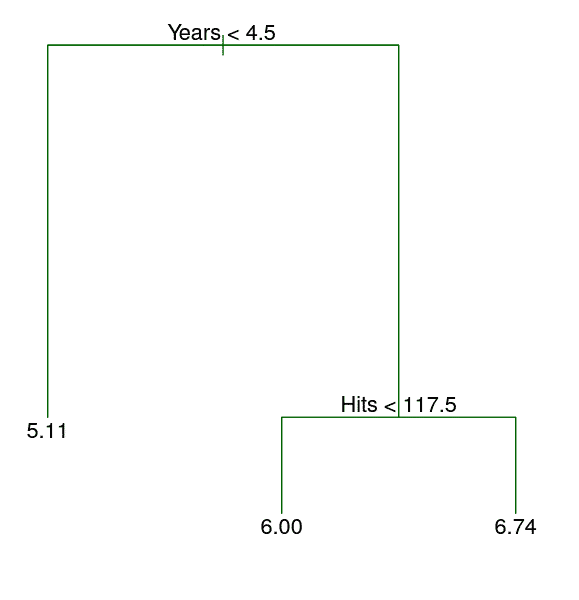
- \(\hat{y}\): 6.74 (logged USD in 1000s)
- Henderson’s salary: 7.42 (logged USD in 1000s)
Single Decision Trees—A Closer Look
Splitting the Feature Space
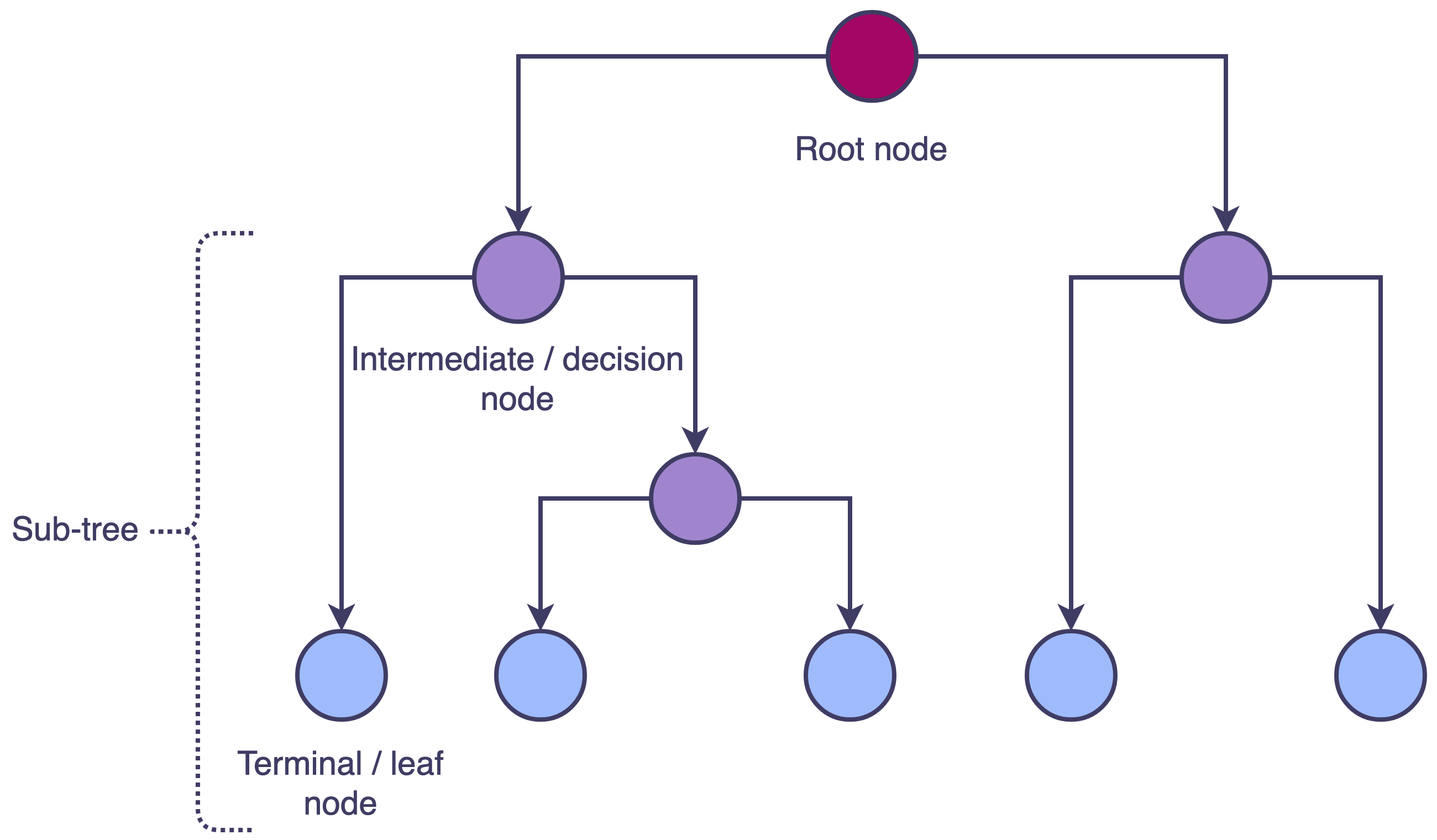
Decision tree estimators rely on recursive binary splitting to segment the feature space.
This procedure begins by selecting the predictor (\(X_j\)) and cutpoint (\(s\), or level of \(X_j\)) such that splitting the feature space into regions {\(X\) | \(X_j\) \(\le\) \(s\)} and {\(X\) | \(X_j\) \(>\) \(s\)} leads to the largest possible reduction in a loss function.
The process is then repeated—i.e., by finding the feature and cutpoint that yields the largest possible reduction in a loss function within the two new regions.
The algorithm generates more splits or branches until a stopping criterion is reached: e.g., tree depth.
Overall, this procedure is top-down and greedy.
Tuning Hyperparameters
Single decision trees are relatively easy to interpret and can account for complex interactions linking \(X\) to \(Y\). However, they are prone to overfitting.
To combat variance across samples, analysts can turn to hyperparameter optimization—i.e., by tuning some of the following hyperparameters:
Maximum Tree Depthrefers to the longest path from the root node to a leaf or terminal node.Minimum Samples for a Splitrefers to the minimum number of training samples that must be present for an internal node to be split further.Minimum Samples for a Leafrefers to the minimum number of training samples that must be present in both branches of a potential split.

Pruning Trees
Beyond tuning a decision tree’s hyperparameters, analysts can combat overfitting by leveraging cost complexity pruning—here are the steps:
- Grow a very large decision tree, \(T_0\).
- Supply different values of the penalty parameter \(\alpha\)—each value corresponding to a less complex subtree \(T \subset T_0\).
- Use cross-validation to find optimal \(\hat{\alpha}\) value.
- Select the subtree \(T \subset T_0\) corresponding to the optimal \(\hat{\alpha}\) value.

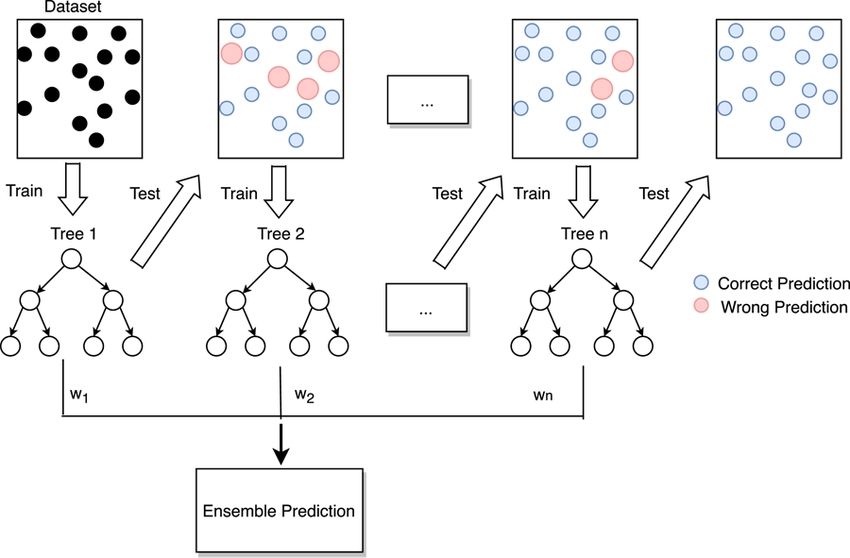
Decision Tree Ensembles
Bagging and Random Forests
If we combine the results of many decision trees—or grow an ensemble of tree-based models—the predictive power of our decision tree estimators can be significantly enhanced.

Image can be retrieved here.
- Bootstrap aggregation—or bagging—is an ensemble learning procedure that is implemented as follows:
- We repeatedly sample from training data with replacement—i.e., bootstrapping—until we have \(B\) synthetic (and independent) datasets with the same dimensions as our training data.
- We grow \(B\) large decision trees on each of the \(B\) (e.g., 1000) datasets.
- We aggregate predictions from each of the \(B\) decision trees (via averaging or voting) to generate our final prediction, \(\hat{f}_{bag}(x)\).
- See Canvas for details on out-of-bag scores for ensemble trees.
Random forest algorithms blend bagging and feature randomness to ensure that decision trees within the ensemble are uncorrelated.
Within each of the \(B\) decision trees, the random forest algorithm only considers a random subset, \(m\), of the \(p\) features (e.g., \(m \approx \sqrt{p}\)) at each decision point.
Like before, we aggregate predictions from each of the \(B\) decision trees (via averaging or voting) to generate our final prediction, \(\hat{f}_{bag}(x)\).
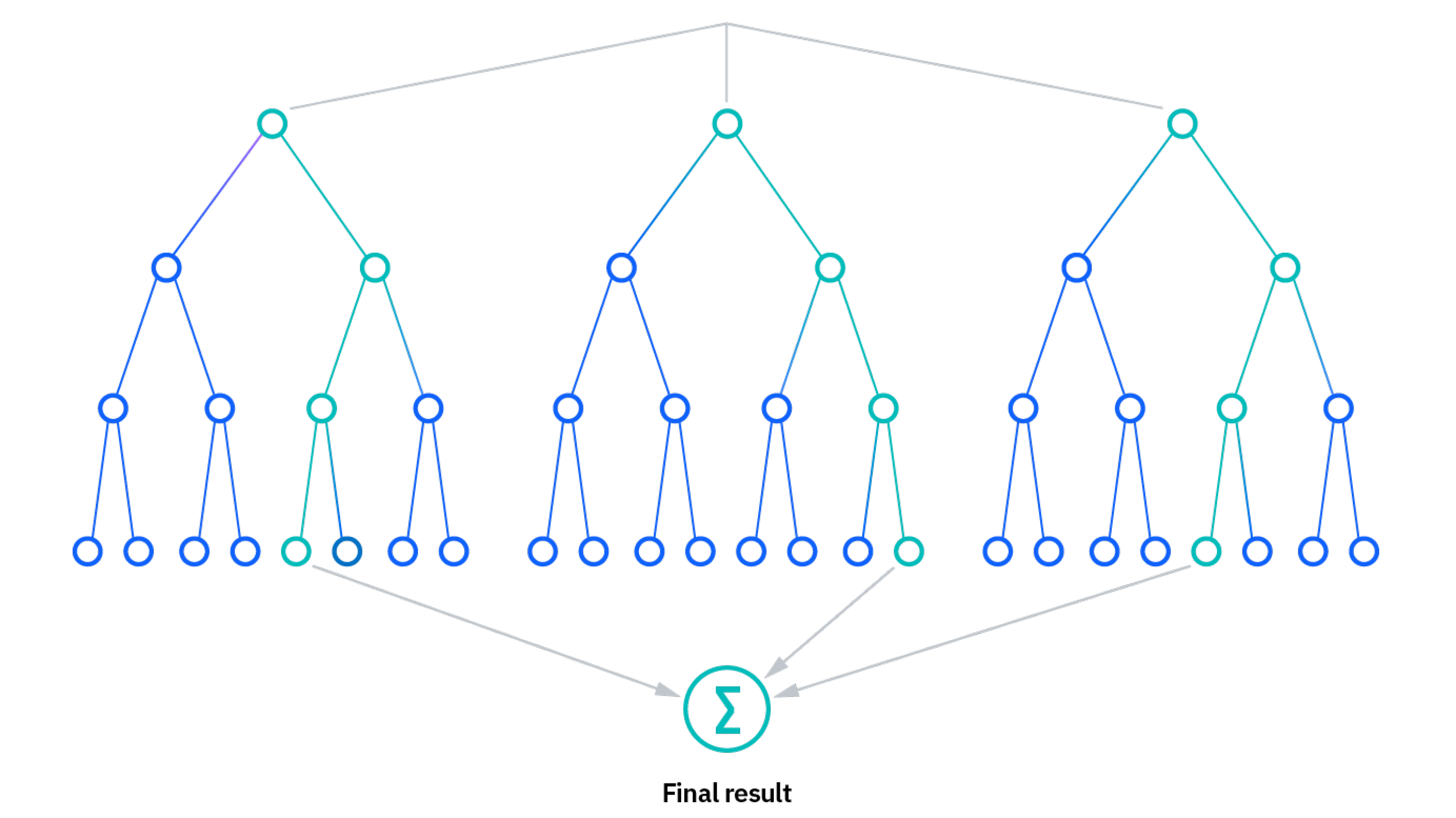
Image can be retrieved here.
Gradient Boosted Trees
Boosting is another approach to ensemble learning—i.e., where small decision trees (weak learners) are sequentially grown to attack the residuals left over by their predecessors.
At each boosting stage (or iteration), we slowly improve \(\hat{f}\) by learning to account for some of the residual errors inherited from previous trees.
For gradient boosted trees, important hyperparameters include \(B\) (the number of trees in the ensemble) and \(\lambda\)—the shrinkage parameter.
Ultimately, predictions are made by passing observations through the entire ensemble of trees (with each tree weighted by \(\lambda\)).
Growing Random Forests
Random Forests in Python
The code snippets embedded below—and their companion Colab Notebook—can be found by clicking here.
Importing Libraries, Loading Data
# Importing pandas to load/wrangle the data, modules from scikit-learn to fit random forest classifier:
import pandas as pd
from sklearn import metrics
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
# Load input data frame:
penguins = pd.read_stata('https://github.com/sakeefkarim/python_plotting/blob/main/files/palmerpenguins.dta?raw=true')
penguins = penguins.dropna()Performing Train-Test Split
# Target:
y = penguins['species']
# Generating input table:
X = penguins.drop(columns = ['species'])
# Adjusting categorical inputs:
adjusted_island = pd.get_dummies(X["island"], drop_first=True)
adjusted_sex = pd.get_dummies(X["sex"], drop_first=True)
# Final set of features:
X = pd.concat([X, adjusted_island, adjusted_sex], axis=1).drop(columns = ['island', 'sex'])
# Train-test split
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify = y, test_size = 0.2, random_state = 416)Fitting Random Forest Classifier
# Developing random forest classifier with 1000 decision trees:
rf = RandomForestClassifier(n_estimators=1000, max_depth = 5, random_state = 416)
# Fitting model:
rf.fit(X_train, y_train)
# Evaluating out-of-sample performance:
rf_pred = rf.predict(X_test)
print(metrics.classification_report(rf_pred, y_test))A Quick Exercise
Using data from {palmerpenguins}, develop a random forest regressor to predict a numeric outcome of interest. Along the way, try to:
- Settle on the optimal number of estimators (
n_estimators) for your classifier using a grid search of possible hyperparameter values for \(B\). - Report your algorithm’s out-of-bag score or out-of-sample performance.
The End